Benchmark functions vs parameter estimation: new paper accepted on Applied Soft Computing
For some real world problems, common algorithms can outperform the most advanced meta-heuristics. This is the case of biochemical parameter estimation (PE), where Particle Swarm Optimization (PSO) and similar algorithms show excellent performances.
We investigated this issue using a bunch of wide-spread meta-heuristics: artificial bee colony (ABC), covariance matrix adaptation evolution strategy (CMA-ES), differential evolution (DE), estimation of distribution algorithm (EDA), genetic algorithms (GA), classic PSO and its settings-free variant Fuzzy Self-Tuning PSO (FST-PSO).
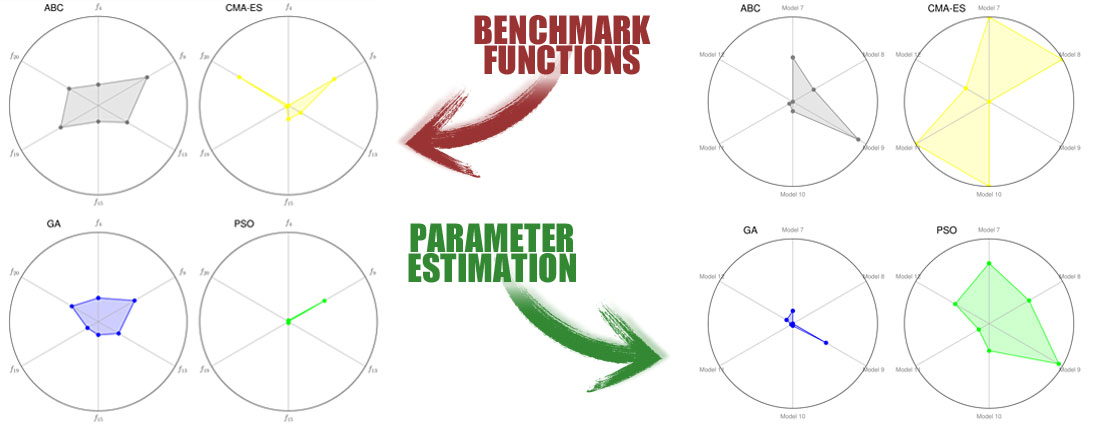
The performance of these 7 meta-heuristics were compared on two test-beds: in the first one, we used multi-modal, shifted/rotated, composed hybrid benchmark functions contained in the CEC’13 suite; in the second one, we performed the PE on multiple synthetic biochemical models on increasing complexity.
The striking results that we collected show that some algorithms working extremely well on benchmark functions can be characterized by considerably poor performances when applied to the PE problem (and vice versa). Moreover, we also highlight that bad performances can be due to the naïve representation of the candidate solutions.
The paper, titled “Biochemical parameter estimation vs. benchmark functions: a comparative study of optimization performance and representation design” was published on Applied Soft Computing.
No Comments