Unsupervised AI and mass-spectroscopy: new paper on Expert Systems with Applications
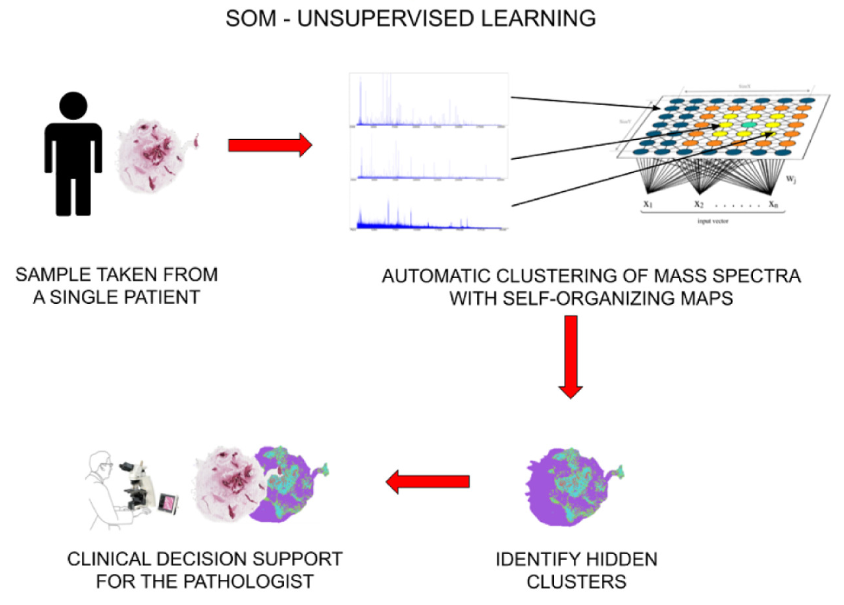
MALDI mass-spectroscopy is a interesting experimental technique that, in a nutshell, breaks down an organic sample into small voxels and, for each voxel, it determines its spectral footprint. In general, given the same tipe of tissue, benign and malignant cells tend to yield different spectra. Since several thousands of spectra can be generated by a single MALDI run, it can be unfeasible to manually review all spectra looking for some uncommon patterns.
In our latest publication on ESwA, we present a novel tool named DSUUL (Discrimination of Spectra Using Unsupervised Learning) which exploits an unsupervised neural network known as Self-Organizing Map (SOM) to automatically separate cells of different type according to their spectral content.

We tested DSUUL on a case study regarding mass spectra data from thyroid nodules. In our results, DSUUL can effectively split the spectra related to benign and malignant cells, guiding the pathologists in the analysis of the sample, notably without the need of any annotated samples. However, it seems that the SOM can be mislead by tissues that are completely healthy or completely malignant. We will investigate on the automatic detection of such circumstance, in order to print a warning message to the human operator overseeing the process.
No Comments