New paper on PLOS Computational Biology
GPUs are basically multi-core co-processors performing vectorial computation applying the so-called SIMD paradigm, i.e., the very same computation (implemented as a kernel) is performed across different data. The computation can be, for instance, a biochemical simulation. In this case, the simulation is defined coarse-grained and it is the case of cupSODA or cuTauLeaping. However, the …
New paper on Scientific Reports
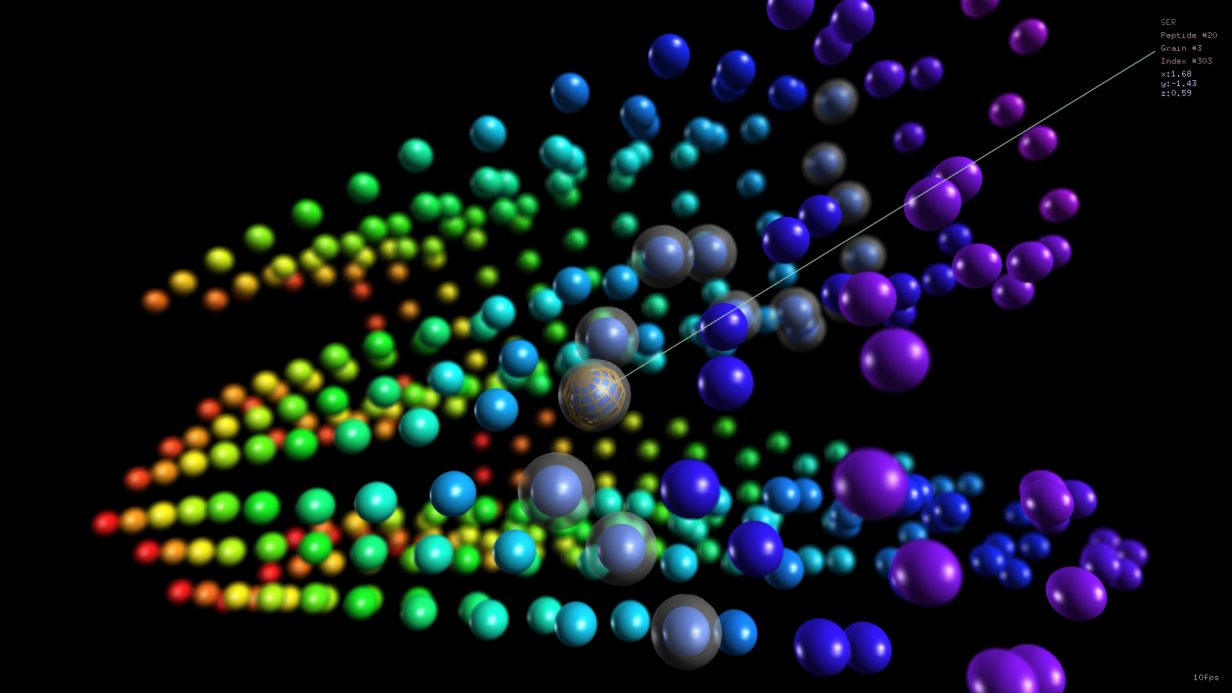
Self-assembling peptides can be useful for clinical applications, for instance as scaffolds based on beta-sheets motifs. The dynamic evolution and self-assemly of peptide systems can be investigated in silico by using molecular dynamics. In particular, coarse-grained simulation exploiting MARTINI force fields can strongly reduce the overall running time and still provide useful information. In a …
New paper: unsupervised learning for medical images analysis
We recently published a new paper on Journal of Supercomputing. The article focuses on an approach that combines automatic feature extraction and unsupervised learning for the analysis of medical images. The idea is to extract (long) vectors of Haralick features for each pixel, and use that information to train a Self-Organizing Map: similar pixels will …
Two new journal papers
Recently, we got two papers accepted for pubblication. The first one was the followup of our CIBCB 2019 paper, published on IEEE Journal of Biomedical and Health Informatics. The paper describes the last developments of our ProCell tool for cell proliferation analysis, whose GUI was completely overhauled by designer Luca Zanini and whose simulation core …


Recent Comments