New paper on Computer Methods and Programs in Biomedicine
Late Gadolinium Enhancement (LGE) is a standard technique for Cardiovascular Magnetic Resonance (CMR), allowing the non-invasive analysis of myocardial issues. The main issue of classic LGE is that blood can appear as bright as scar regions, which complicates the interpretation of results. In the last decade, multiple “Dark-Blood” LGE (DB-LGE) approaches have been proposed to …
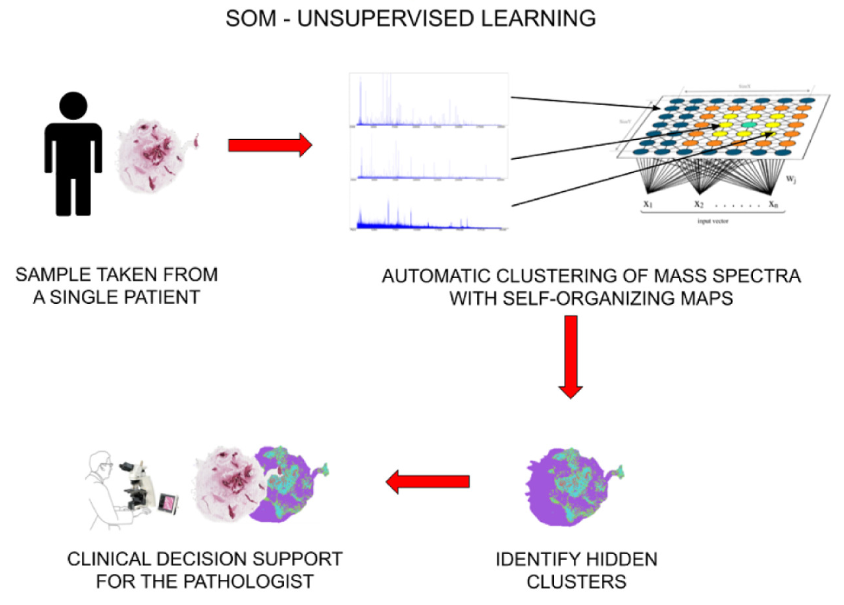
Unsupervised AI and mass-spectroscopy: new paper on Expert Systems with Applications
MALDI mass-spectroscopy is a interesting experimental technique that, in a nutshell, breaks down an organic sample into small voxels and, for each voxel, it determines its spectral footprint. In general, given the same tipe of tissue, benign and malignant cells tend to yield different spectra. Since several thousands of spectra can be generated by a …
Special Issue on IEEE Computational Intelligence Magazine
Computational Intelligence (CI) provides a set of powerful tools to effectively tackle complex computational tasks: global optimization methods (e.g., evolutionary computation, swarm intelligence), machine learning (e.g., neural networks), and fuzzy reasoning. While CI research activity generally focuses on the improvement of the algorithms (e.g., faster convergence, higher accuracy, reduced error), there is another promising research …
New paper published on Symmetry
One of the core topics of our research group is mechanistic biochemical modeling. The analysis of such models is often performed using simulations, which can be extreme from a computational point of view. Hence, we worked a lot on the acceleration of methods. In order to investigate the performances our algorithms, we need a large …
New paper on PLOS Computational Biology
GPUs are basically multi-core co-processors performing vectorial computation applying the so-called SIMD paradigm, i.e., the very same computation (implemented as a kernel) is performed across different data. The computation can be, for instance, a biochemical simulation. In this case, the simulation is defined coarse-grained and it is the case of cupSODA or cuTauLeaping. However, the …
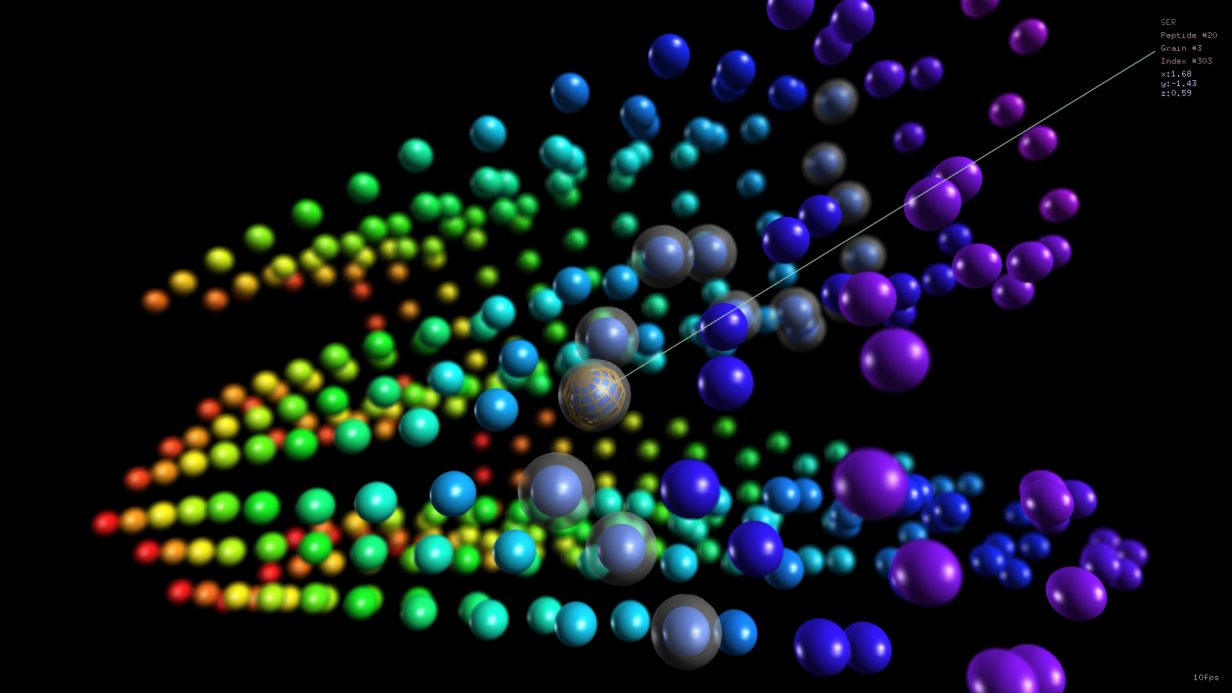
New paper on Scientific Reports
Self-assembling peptides can be useful for clinical applications, for instance as scaffolds based on beta-sheets motifs. The dynamic evolution and self-assemly of peptide systems can be investigated in silico by using molecular dynamics. In particular, coarse-grained simulation exploiting MARTINI force fields can strongly reduce the overall running time and still provide useful information. In a …
New paper: unsupervised learning for medical images analysis
We recently published a new paper on Journal of Supercomputing. The article focuses on an approach that combines automatic feature extraction and unsupervised learning for the analysis of medical images. The idea is to extract (long) vectors of Haralick features for each pixel, and use that information to train a Self-Organizing Map: similar pixels will …
Simpful’s paper accepted
Simpful is now mature enough to have an official paper associated to the project. Our manuscript — describing both Simpful functionalities and inner machinery — was accepted for publication on the International Journal of Computational Intelligence Systems. In the meanwhile, the development of Simpful continues. We will introduce support for additional reasoning systems and, in …
Two new journal papers
Recently, we got two papers accepted for pubblication. The first one was the followup of our CIBCB 2019 paper, published on IEEE Journal of Biomedical and Health Informatics. The paper describes the last developments of our ProCell tool for cell proliferation analysis, whose GUI was completely overhauled by designer Luca Zanini and whose simulation core …





Recent Comments