Roundtable “HAI presente l’AI?”
A few days ago I partecipated to a roundtable focused on the AI Act organized by the PInK office of technology transfer of Ca’ Foscari. The AI Act was approved on May and will become official on August 2024. The regulation covers many aspects and it is particularly relevant in the case of high risk …
Dual seminar on Interpretability at the Jheronimus Academy of Data Science (JADS)
On May 7th, Marie Curie fellow Chiara Gallese and I have been kindly invited by Prof. Uzay Kaymak to give two seminary at the Jheronimus Academy of Data Science (JADS) in ‘s-Hertogenbosch, The Netherlands. Since Chiara’s MSCA revolves around fairness of AI in biomedicine and healthcare, with a focus on non-discriminatory data use and reuse, …
“Shark Tank” award at CMR2024
We won the “Shark Tank” award at CMR2024! THAITI, our AI-powered tool for high precision cardiac magnetic resonance, tailored to the patient, was judged innovative, clinically impactful, with translational value, and commercially-viable by a multidisciplinary team of experts. THAITI was developed together with Dr. Camilla Torlasco, M.D. (cardiologist at the Istituto Auxologico Italiano, Milan, Italy), …
PhD course on Computational Intelligence
From Sep 11 to Sep 15, 2023 I will give five lectures on Computational Intelligence within the “Knowledge, Interaction and Intelligent Systems” PhD course. The focus will be on two main topics: advanced meta-heuristics for global optimization and intepretable AI by means of fuzzy reasoning. Specifically, these are the contents that will be presented: The …
New paper on Computer Methods and Programs in Biomedicine
Late Gadolinium Enhancement (LGE) is a standard technique for Cardiovascular Magnetic Resonance (CMR), allowing the non-invasive analysis of myocardial issues. The main issue of classic LGE is that blood can appear as bright as scar regions, which complicates the interpretation of results. In the last decade, multiple “Dark-Blood” LGE (DB-LGE) approaches have been proposed to …
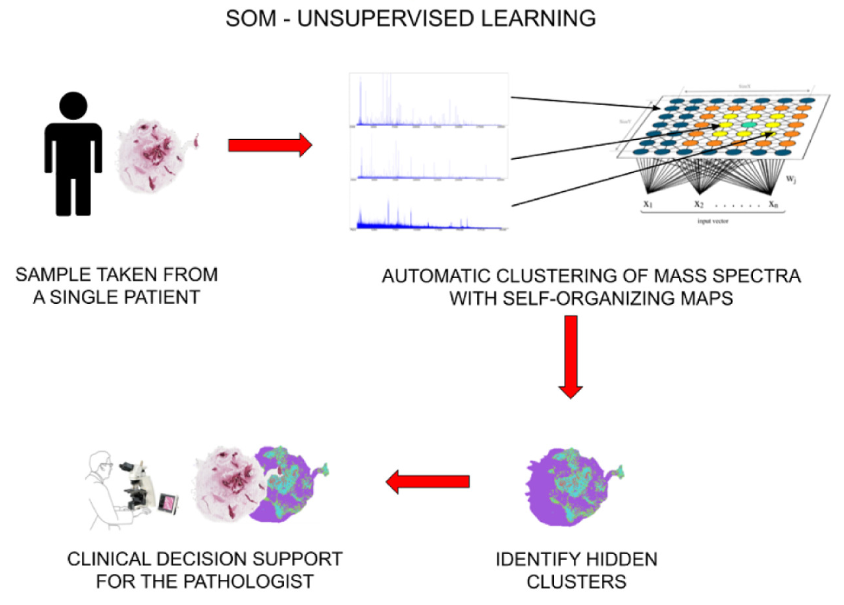
Unsupervised AI and mass-spectroscopy: new paper on Expert Systems with Applications
MALDI mass-spectroscopy is a interesting experimental technique that, in a nutshell, breaks down an organic sample into small voxels and, for each voxel, it determines its spectral footprint. In general, given the same tipe of tissue, benign and malignant cells tend to yield different spectra. Since several thousands of spectra can be generated by a …
Best paper award at IEEE CIBCB 2022
Our manuscript “Predicting and Characterizing Legal Claims of Hospitals with Computational Intelligence: the Legal and Ethical Implications” was awarded as best paper at the 2022 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (IEEE CIBCB). The manuscript investigates the NHS data to determine under which circumstances the COVID pandemics caused an increment of …
Special Issue on IEEE Computational Intelligence Magazine
Computational Intelligence (CI) provides a set of powerful tools to effectively tackle complex computational tasks: global optimization methods (e.g., evolutionary computation, swarm intelligence), machine learning (e.g., neural networks), and fuzzy reasoning. While CI research activity generally focuses on the improvement of the algorithms (e.g., faster convergence, higher accuracy, reduced error), there is another promising research …
New paper published on Symmetry
One of the core topics of our research group is mechanistic biochemical modeling. The analysis of such models is often performed using simulations, which can be extreme from a computational point of view. Hence, we worked a lot on the acceleration of methods. In order to investigate the performances our algorithms, we need a large …
Simone Spolaor recipient of EUSFLAT 2020 best PhD thesis award
My former PhD student, Simone Spolaor, was awarded by EUSFLAT for the best 2020 PhD thesis on fuzzy logic. Prof. Daniela Besozzi (Simone’s main advisor) and me are incredibly proud of this achievement. Simone’s thesis, titled “Fuzzy Logic for the modeling and simulation of complex systems”, is an investigation about moving back and forth from …






Recent Comments